
- Serial cloner feature import how to#
- Serial cloner feature import serial#
- Serial cloner feature import update#
- Serial cloner feature import software#
Serial cloner feature import update#
It also allows to generate sense and antisense sequences to be obtained after bi-sulfite conversion.Preface The Appliance Concept Use Cases Contact 1 Overview 1.1 Basic Workflow 1.2 Conceptual Overview 1.3 Terminology 1.4 System Requirements 2 Installation 2.1 Installation from OBS 2.2 Installation from Distribution Repositories 2.3 Example Appliance Descriptions 3 Quick Start 3.1 Before you start 3.2 Choose a First Image 3.3 Build your First Image 3.4 Run your Image 3.5 Tweak and Customize your Image 4 Working from the Command Line 4.1 kiwi-ng 4.2 kiwi-ng result list 4.3 kiwi-ng result bundle 4.4 kiwi-ng system prepare 4.5 kiwi-ng system update 4.6 kiwi-ng system build 4.7 kiwi-ng system create 4.8 kiwi-ng image resize 4.9 kiwi-ng image info 5 Troubleshooting 5.1 Build Host Constraints 5.2 Architectures 5.3 Host Security Settings Conflicts with KIWI 5.4 Incompatible Filesystem Settings on Host vs.
Serial cloner feature import serial#
Serial Cloner 2.5 now allows to import codon frequency tables from the internet using a Web interface. It is also posible to send directly BLAST request at the NCBI and obtain the result inside a Web interface.
Serial cloner feature import how to#
You will also find a Restriction Enzyme library management interface, Additional tools, like a web browser for direct import of NCBI and EMBL entries, a virtual cutter to prepare restriction analysis or a silent restriction map generator to find how to introduce restriction sites without modifying the translated peptide are also provided. Features are now visible when aligning locally. Finally, Serial Cloner provides an interface to align two sequences using a local algorithm or the BLAST2Seq NCBI server. An additional interface allows easy Gateway(tm) cloning for both BP and LR reactions. Just select, blunt if you need, and click the Ligate button. Finally, you can assemble fragments, obtained by PCR, adaptor/shRNA synthesis or simply by graphically selecting fragments between restriction sites. shRNA constructions based on pre-defined scaffolds are also automated. In addition to the classical restriction maps - both graphic and text-based - or site usage windows, you will be able to quickly extract a sub-sequence either in a selection or between restriction sites, to create a new PCR-based fragment or synthetic adaptors. Serial Cloner will assist you in setting-up new sub-cloning projects and in preparing the electronic versions of your constructs. Version 2.5 now allows to open protein sequences, align protein sequences and reverse translate proteins using defined codon biais tables. The Collection of Features to be scanned for can be defined and modified by the user, imported and exported. All the features are displayed on the map using user-modifiable colors the features can be manually entered, imported from GenBank or automatically found after scanning by Serial Cloner. The graphic map of Serial Cloner is really Graphic as you can easily select and extract a fragment or show single, double or multiple cutter all in the same window. Since version 2.1, Sequence Features are visible in the Sequence Map. Serial Cloner also lets you build text restriction map and quickly format it to add multi-frame translation or only show single cutters for example.
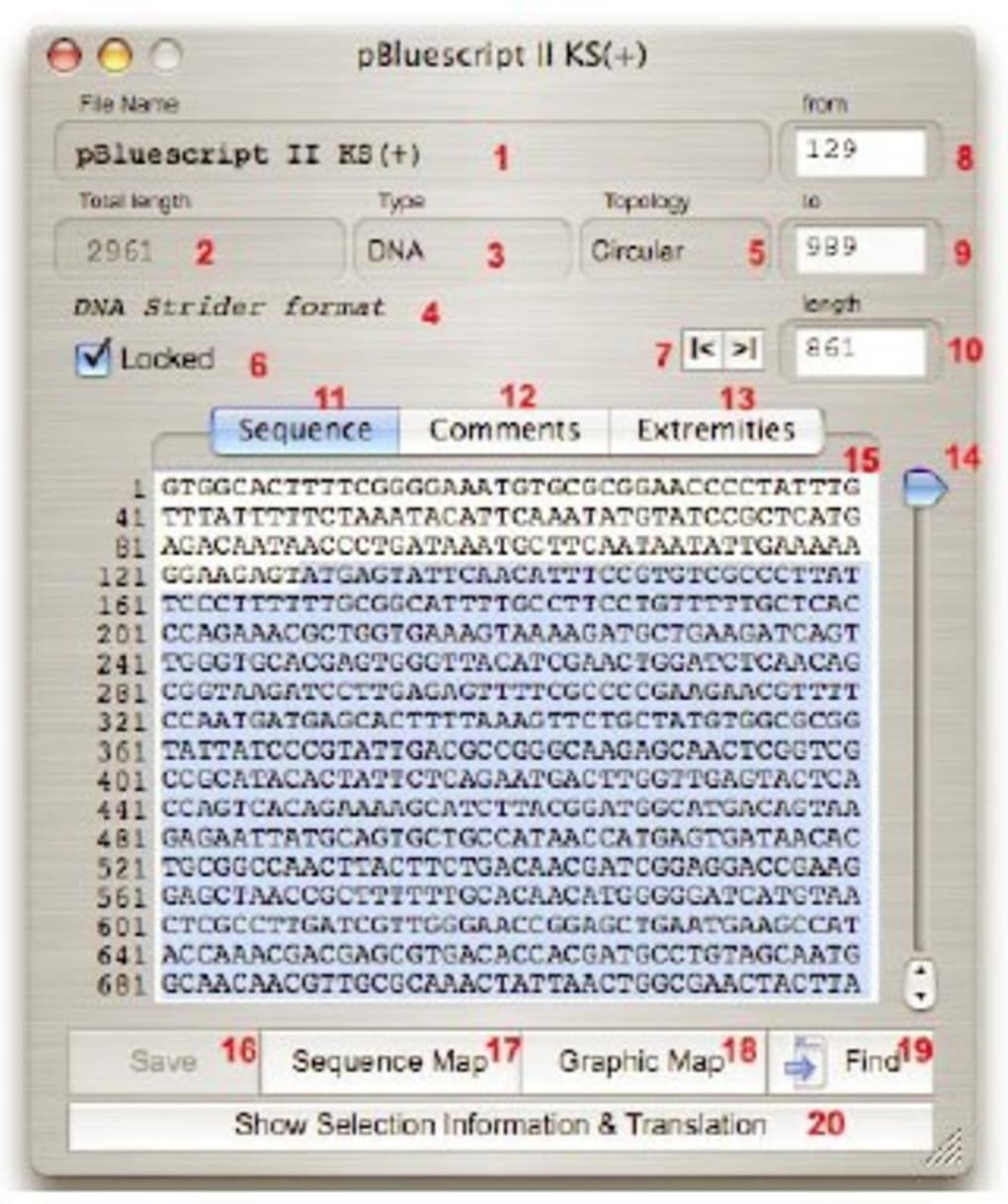
Numerically select fragments, find restriction sites, ORF or any nucleotide or peptide sequence, calculate Tm of selected fragments, %GC or dynamically determine the translation your selection into peptide and calculate the MW using a compact interface. Serial Cloner 2.5, handles Annotations and Features both in the sequence and in the Graphic Map and can automatically scan for sequence Features.Īll the tools you need to analyze and manipulate your sequences are available in an all-in-one-window concept. Powerful graphical display tools and simple interfaces help the analysis and construction steps in a very intuitive way. Import from VectorNTI multi-file format is also supported. Serial Cloner also import files saved in the Vector NTI, MacVector, ApE, DNAstar, pDRAW32 and GenBank formats. Serial Cloner reads and write DNA Strider-compatible files and import and export files in the universal FASTA format.
Serial cloner feature import software#
Serial Cloner has been developed to provide a light yet powerful molecular biology software to both Macintosh and Windows users. Serial Cloner has been developed to provide a light yet powerful molecular biology software to its users.


 0 kommentar(er)
0 kommentar(er)
